指环指泛素 E3 连结基因 TaSDIR1-4A 有助于确定普通小麦的粒状大小
 Journal of Experimental Botany, Vol. 71, No. 18 pp. 5377–5388, 2020 doi:10.1093/jxb/eraa271 Advance Access Publication 1 June 2020
Journal of Experimental Botany, Vol. 71, No. 18 pp. 5377–5388, 2020 doi:10.1093/jxb/eraa271 Advance Access Publication 1 June 2020
This paper is available online free of all access charges (see https://academic.oup.com/jxb/pages/openaccess for further details)
RESEARCH PAPER
![]() RING finger ubiquitin E3 ligase gene TaSDIR1-4A contributes to determination of grain size in common wheat
RING finger ubiquitin E3 ligase gene TaSDIR1-4A contributes to determination of grain size in common wheat
![]()
![]()
![]() Jingyi Wang1,†, , Ruitong Wang1,†, Xinguo Mao1, Jialing Zhang1, Yanna Liu1, Qi Xie2, Xiaoyuan Yang2, Xiaoping Chang1, Chaonan Li1, Xueyong Zhang1, and Ruilian Jing1,*,
Jingyi Wang1,†, , Ruitong Wang1,†, Xinguo Mao1, Jialing Zhang1, Yanna Liu1, Qi Xie2, Xiaoyuan Yang2, Xiaoping Chang1, Chaonan Li1, Xueyong Zhang1, and Ruilian Jing1,*,
1 National Key Facility for Crop Gene Resources and Genetic Improvement/Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing 100081, China
2 Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China
† These authors contributed equally to this work.
* Correspondence: jingruilian@caas.cn
Received 25 November 2019; Editorial decision 22 May 2020; Accepted 26 May 2020 Editor: Dabing Zhang, Shanghai Jiao Tong University, China
Abstract
Salt and drought-induced RING finger1 (SDIR1) is a RING-type E3 ubiquitin ligase that plays a key role in ABA- mediated responses to salinity and drought stress via the ubiquitination pathway in some plant species. However, its function in wheat (Triticum aestivum) is unknown. Here, we isolated a SDIR1 member in wheat, TaSDIR1-4A, and characterized its E3 ubiquitin ligase activity. DNA polymorphism assays showed the presence of two nucleo- tide variation sites in the promoter region of TaSDIR1-4A, leading to the detection of the haplotypes Hap-4A-1 and Hap-4A-2 in wheat populations. Association analysis showed that TaSDIR1-4A haplotypes were associated with 1000-grain weight (TGW) across a variety of different environments, including well-watered and heat-stress condi- tions. Genotypes with Hap-4A-2 had higher TGW than those with Hap-4A-1. Phenotypes in both gene-silenced wheat and transgenic Arabidopsis showed that TaSDIR1-4A was a negative regulator of grain size. Gene expression assays indicated that TaSDIR1-4A was most highly expressed in flag leaves, and expression was higher in Hap-4A-1 acces- sions than in Hap-4A-2 accessions. The difference might be attributable to the fact that TaERF3 (ethylene response factor) can act as a transcriptional repressor of TaSDIR1-4A in Hap-4A-2 but not in Hap-4A-1. Examination of modern wheat varieties shows that the favorable haplotype has been positively selected in breeding programs in China. The functional marker for TaSDIR1-4A developed in this study should be helpful for future wheat breeding.
Keywords: Association analysis, functional marker, RING E3 ubiquitin ligase, TGW, Triticum aestivum, VIGS, wheat.
![]()
Introduction
Given rapid increases in population together with climate change, we could soon witness global food shortages (Boyer, 1982; Godfray et al., 2010; Tilman et al., 2011). Grains are the most important food sources for humans (Zuo and Li, 2014; Li et al., 2018), and grain size is an important com- ponent of grain weight. Therefore, an understanding of the mechanisms underlying seed development is important for
improvement of grain yield in crop species. Recent studies have shown that ubiquitin-proteasome pathways are in- volved in control of seed size (Li and Li, 2016; Nadolska- Orczyk et al., 2017; Kelley, 2018; Li et al., 2019b; Xu and Xue, 2019), and one of these, the DA1 pathway, plays a key role in controlling seed size in Arabidopsis by regulating cell proliferation in the integuments (Disch et al., 2006; Xia
![]()
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Experimental Biology.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted reuse, distribution, and reproduction in any medium, provided the original work is properly cited.
![]()
et al., 2013a; Du et al., 2014; Li and Li, 2016; Dong et al., 2017). This pathway of control of seed size is highly con- served among species (Su et al., 2011; Wang et al., 2017; Xie et al., 2018). For example, Grain Width and Weight2 (GW2), a major quantitative trait locus (QTL) in rice, is a homolog of DA2, a RING-type E3 ubiquitin ligase gene (Song et al., 2007; Xia et al., 2013a). GW2 homologs in maize and wheat are also involved in the control of grain size (Li et al., 2010b; Bednarek et al., 2012; Zhang et al., 2018).
Another RING-type E3 ubiquitin ligase, salt and drought- induced RING finger1 (SDIR1), has been shown to primarily participate in stress responses in numerous species, including Arabidopsis (Zhang et al., 2007, 2015), maize (Xia et al., 2012; Liu et al., 2013), rice (Gao et al., 2011), tobacco (Xia et al., 2013b), and grape (Tak and Mhatre, 2013), while others have been identified to not only be involved in stress responses, but also in plant growth and grain development (Sun et al., 2019). In wheat, there are two AtSDIR1 homologs,TaSDIR1-4A and TaSDIR1-4D, and the latter might be involved in abiotic stress responses (Wang et al., 2018). To date, the role of TaSDIR1 in grain size development in wheat is unknown.
Based on gene-sequence polymorphisms, candidate gene as- sociation analysis has been shown to be an efficient approach to reveal relationships between gene/polymorphic loci and traits, and it has been widely used in many crop species, including maize (Wang et al., 2016b) and wheat (Zhang et al., 2017). A series of elite alleles and molecular markers associated with important traits have been discovered using this method (Cao et al., 2020), and pyramiding these elite alleles through marker- assisted selection (MAS) will greatly accelerate the process of wheat breeding. Hence, finding elite alleles associated with im- portant agronomic traits is considered as being fundamental to wheat genetic improvement.
In the present research, we identified the RING-type E3 ubiquitin ligase gene TaSDIR1-4A and performed association analysis using multiple wheat populations, which demonstrated that it was significantly associated with 1000-grain weight.The favorable haplotype/allele has been positively selected for in Chinese wheat breeding programs, and the functional marker that we have developed for this haplotype will be beneficial in future programs.
Materials and methods
Plant materials
The common wheat (Triticum aestivum) cultivar Hanxuan 10 (H10) was used for cloning and structural analysis of the TaSDIR1 genes. A diverse wheat population of 32 accessions previously screened with 209 SSR markers (Zhang et al., 2013) was used for gene sequencing and detection of nucleotide polymorphisms (Supplementary Table S1 at JXB online).
Four wheat populations were used in this study, as follows. Population 1 was a doubled-haploid (DH) population with 150 lines developed from the cross H10 × Lumai 14 (L14) and was used for genetic mapping. H10 was selected from more than 20 000 wheat accessions due to its strong drought tolerance, whilst L14 is a high-yielding cultivar adapted to well- watered and fertile conditions, and was widely grown in northern China during the 1990s. Population 2 (262 accessions) was used for association analysis and consisted of 209 modern varieties, 43 advanced lines, and 10 landraces (Zhang et al., 2013). Population 3 comprised 157 landraces, and
Population 4 comprised 348 modern cultivars mainly from a Chinese wheat mini-core collection that represents more than 70% of the genetic diversity of the entire Chinese germplasm collection (Hao et al., 2011). Populations 3 and 4 were also used for determination of haplotype fre- quencies in 10 different wheat production zones covering the whole of China (Zhang et al., 2002).
Growth conditions and measurement of agronomic traits
![]() Population 1 was planted in 10 environments (year×site×water re- gime combinations) at Shunyi (40°230′N; 116°560′E) and Changping (40°130′N; 116°130′E) in Beijing during the 2015–2016 and 2016– 2017 growing seasons. Population 2 was planted in 10 environments (year×site×water regime×heat stress combinations), including the same sites as Population 1, during 2010–2011, 2011–2012, and 2012–2013 for measurement of the following agronomic traits: plant height (PH), spike length (SL), peduncle length (PLE), length of penultimate node (LPN), number of spikes per plant (NSP), number of spikelets per spike (NSS), number of grains per spike (NGS), and 1000-grain weight (TGW). Two water regimes were applied at each site, namely well-watered (WW) and rain-fed (drought stress, DS). The WW plots were irrigated with 750 m3 ha–1 (75 mm) water at pre-overwintering, jointing, flowering, and grain-filling, whereas the DS plots were rain-fed only. The rainfalls were 131 mm in the 2010–2011 growing season, 180 mm in 2011–2012,
Population 1 was planted in 10 environments (year×site×water re- gime combinations) at Shunyi (40°230′N; 116°560′E) and Changping (40°130′N; 116°130′E) in Beijing during the 2015–2016 and 2016– 2017 growing seasons. Population 2 was planted in 10 environments (year×site×water regime×heat stress combinations), including the same sites as Population 1, during 2010–2011, 2011–2012, and 2012–2013 for measurement of the following agronomic traits: plant height (PH), spike length (SL), peduncle length (PLE), length of penultimate node (LPN), number of spikes per plant (NSP), number of spikelets per spike (NSS), number of grains per spike (NGS), and 1000-grain weight (TGW). Two water regimes were applied at each site, namely well-watered (WW) and rain-fed (drought stress, DS). The WW plots were irrigated with 750 m3 ha–1 (75 mm) water at pre-overwintering, jointing, flowering, and grain-filling, whereas the DS plots were rain-fed only. The rainfalls were 131 mm in the 2010–2011 growing season, 180 mm in 2011–2012,
158 mm in 2012–2013, 161 mm in 2015–2016, and 173 mm in 2016–
2017. A heat-stress (HS) treatment was applied 1 week post-anthesis at Shunyi by positioning a plastic film supported by steel frames over the plots in the field (Li et al., 2019a). Populations 3 and 4 were planted in three environments, at Luoyang (34°610′N; 112°450′E) in Henan prov- ince in 2002 and 2005, and at Shunyi in 2010. In all cases, each accession was planted in a 2-m, 4-row plot with a row spacing of 30 cm and 40 seeds per row. Five plants in the middle of each plot were sampled for phenotyping at maturity.
Isolation of TaSDIR1-4A and construction of a phylogenetic tree
The reference sequence of TaSDIR1-4A was obtained from the URGI website (https://wheat-urgi.versailles.inra.fr/). Genome-specific pri- mers were designed based on the gene sequence (all primers are listed in Supplementary Table S2). Genomic DNA and cDNA from cv. H10 were used as templates. The gene structure of TaSDIR1-4A was determined using Lasergene 7.1.0 (DNASTAR, Inc., Madison, WI, USA) by align- ment of the amplified cDNA and genomic DNA sequences.A neighbor- joining phylogenetic tree was constructed using the MEGA software v5.2 (https://www.megasoftware.net/).
E3 ubiquitin ligase activity assays
The full-length TaSDIR1-4A ORF (840 bp) was cloned into a pMAL- C5X vector that has a maltose-binding protein (MBP) tag through seamless DNA cloning using In-Fusion® HD Cloning Plus (638910, TaKaRa). A TaSDIR1-4A mutant (His-244 to Tyr) containing a muta- tion in the RING finger domain was constructed using a Site-directed Mutagenesis Kit (B639281, Sangon Biotech, Beijing) according to the manufacturer’s protocol. MBP, MBP-AtSDIR1, MBP-TaSDIR1-4A, and MBP-TaSDIR1-4AH244Y were prepared and purified in Escherichia coli. MBP was used as a negative control, and MBP-AtSDIR1 was the positive control. In vitro E3 ligase assays were performed as described by Xie et al. (2002). Briefly, E1 (from wheat, 50 ng), E2 (UBCh5b, 100 ng), E3 (1μg), and 6×His tag ubiquitin (Ub, 4 μg) were mixed and incubated at 30 °C for 60 min. The mixture was separated by SDS-PAGE and blotted onto PVDF membranes (Millipore, IPVH00010). Anti-Ub and anti-MBP antibodies were used and bands were detected using a Thermo Pierce ECL (NCI4106) according to the manufacturer’s instructions.
Sequence polymorphisms in TaSDIR1-4A
Nucleotide polymorphisms in TaSDIR1-4A sequences were detected by scanning a small diverse panel of 32 common wheat accessions.
Genomic DNA was extracted from young leaves of 10-d-old seed- lings using the CTAB method. TaSDIR1-4A fragments were amplified using primers specific to the A genome. Target bands were purified and cloned into the pEASY-Blunt vector, and transformed into Trans1-T1 phage-resistant chemically competent cells (CD501-03, Beijing Trans Gen Biotech Co., Ltd.) using the heat-shock method. Twelve positive clones were randomly selected for sequencing using a 3730 XI DNA Analyzer (ABI). TaSDIR1-4A nucleotide se- quence polymorphisms were obtained using the SeqMan program in Lasergene 7.10 (DNASTAR).
Functional marker development
A functional marker was developed based on a SNP (G/A) at position
–395 bp, by mismatching bases in the dCAPS primer to create a re- striction site. The first-round PCR product obtained by using primers specific to the A genome was used as a template for the second round of PCR, which was performed as follows: 95 °C for 5 min, followed by 33 cycles of 95 °C for 30 s, annealing 58 °C for 30 s, and extension at 72 °C for 30 s, with a final extension of 72 °C for 10 min. The PCR products were digested with restriction enzyme at 37 °C for 2 h and then separ- ated by electrophoresis in 4% agarose gels.
Chromosome location and linkage analysis
Diploid and tetraploid wheat relatives with various genomes and a set of nullisomic-tetrasomic lines of the common wheat cultivar Chinese Spring were used to identify the genomic origin and chromosome location of TaSDIR1-4A.The H10 × L14 DH population, and the IciMapping V4.1 software (www.isbreeding.net/software/) were used for gene mapping and linkage analysis.
Population structure and association analysis
The population structure of Population 2 has previously been scrutinized with STRUCTURE v2.3.4 using data from 209 SSR markers evenly distributed on the 21 chromosomes in wheat (Li et al., 2012b). The two subpopulations comprised 110 and 152 accessions. Association mapping was performed using the mixed linear model in TASSEL v2.1 (https:// www.maizegenetics.net/tassel), which accounted for population struc- ture. One-way ANOVA was performed in SPSS 19.0 to identify signifi- cant associations between gene haplotypes and agronomic traits. Tukey tests were used to determine significant differences at P=0.05.
Virus-induced gene silencing (VIGS)
A series of recombinant barley stripe mosaic virus (BSMV) vectors was constructed.A non-conserved TaSDIR1-4A fragment of 253 bp spanning the 3´-terminal sequence and 3´-UTR region was selected to construct the BSMV vector BSMV:TaSDIR1-4A.This target region was compared with the Chinese Spring genome assembly v1.0 sequence (http://plants. ensembl.org/Triticum_aestivum/Tools/Blast) to ensure its specificity. A BSMV vector containing the GFP sequence was used as a control. To construct recombinant BSMV, the BSMV-γ vector was digested at the Not I restriction site, and the TaSDIR1-4A fragment amplified from the cDNA of Chinese Spring was inserted into the vector by seamless DNA cloning. The BSMV was inoculated onto leaves of jointing wheat plants following the procedures described in Wei et al. (2019). Flag leaves from BSMV-infected plants were collected at anthesis for determination of gene expression patterns. At maturity, seeds were collected, dried, and kernel size and weight were determined on samples of at least 200 seeds using an automatic seed measurement device (SC-A1, WSeen). Results were obtained from independent experiments.
Production of transgenic Arabidopsis TaSDIR1-4A-overexpression lines
Seeds of the sdir1-1 (SALK_052702) mutant were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/. The
full-length cDNA of TaSDIR1-4A with the 35S promoter was inserted into the modified pCAMBIA1300 vector at the Xba I and Kpn I restriction en- zyme sites. Agrobacterium harboring TaSDIR1-4A and empty vectors were transferred into wild-type and sdir1-1 Arabidopsis flowers using the floral dip method to obtain transgenic lines (Clough and Bent, 1998). The seed sizes of homozygous T3 generation plants were measured using a SC-A1 device.
Expression of TaSDIR1-4A in wheat
![]() The H10 cultivar was used for expression pattern analysis of TaSDIR1-4A. At the flowering stage, spikes, flag leaves, stems, nodes, root tissue at the stem base, and root tissues at different depths were sampled for spatial- temporal analysis of expression patterns (Wang et al., 2019).
The H10 cultivar was used for expression pattern analysis of TaSDIR1-4A. At the flowering stage, spikes, flag leaves, stems, nodes, root tissue at the stem base, and root tissues at different depths were sampled for spatial- temporal analysis of expression patterns (Wang et al., 2019).
Twelve accessions of each of the two different haplotypes were ran- domly selected from each of Populations 1 and 2 (Supplementary Table S3). Flag leaves at anthesis were sampled from field-grown plants and used to assess the expression levels.
Based on pilot experiments, 2-week-old wheat seedlings were sprayed with 50 μM ABA solution or treated at 250 mM NaCl for 0.5–72 h. Whole plants were then sampled for detection of stress-induced expres- sion of TaSDIR1-4A.
Yeast one-hybrid assays
Yeast one-hybrid assays were performed to check the binding of the ethylene response factor DNA binding domain (TaERF-BD) to the TaSDIR1-4A promoter region. The pB42AD vector was digested with restriction enzyme Not I in order to insert the TaERF-BD. The TaERF-BD fragment was then cloned into the vector through seamless DNA cloning. Six fragments of the TaSDIR1-4A promoter region were cloned into pLacZi as the reporter gene plasmid as follows. For the H10 construct, a 588-bp (–757 to –170 bp) fragment from the TaSDIR1-4A promoter region of H10 (Hap-4A-1) was amplified and cloned into pLacZi by seamless DNA cloning. For the L14 construct, a 586-bp (–755 to –170 bp) fragment (Hap-4A-2) was amplified and cloned using the same method. For the ACC-H10 Box construct, the target frag- ment contained the triplicates of the ACC Box (32 bp, –418 to –387 bp from H10). For the GCC-H10 Box construct, the same fragment with an A-to-G mutation was used. For the GCC-L14 Box construct, the target fragment contained the triplicates of the GCC Box (32 bp, –418 to
–387 bp from L14). For the ACC-L14 box construct, the same fragment with a G-to-A mutation was used. These four vectors were separately cloned into pLacZi by whole-gene synthesis at the Beijing Genomics Institute (BGI). Plasmids for the pB42AD vector with the TaERF-BD were co-transformed with the six reporter gene constructs into yeast strain EGY48 using standard yeast transformation methods (Lin et al., 2007).The yeast was incubated on media lacking Ura and Leu with x-α- gal. Interactions between TaERF-BD and each promoter fragment were tested by LacZ staining (Wang et al., 2016a).
Purification of the TaERF-BD protein and electrophoretic mobility shift assays (EMSAs)
TaERF-BD cDNA (219 bp) was fused into the pGEX-4T1 vector, which has a glutathione S-transferase (GST) tag, using seamless DNA cloning at an EcoR I site using In-Fusion® HD Cloning Plus. TaERF-BD protein expression in Escherichia coli BL21 cells was induced by 0.5 mM isopropyl- β-d-thiogalactoside (IPTG) at 28 °C for 6 h, and it was purified using glutathione–Sepharose 4B (52-2303-00, GE Healthcare). Unlabeled GCC-L14 and ACC-L14 Boxes, biotin-labelled GCC-L14 and ACC- L14, and GCC-H10 and ACC-H10 Boxes and their reverse comple- mentary sequences were synthesized and annealed as probes. EMSA was carried out using a LightShift® Chemiluminescent EMSA Kit (20148, Thermo Scientific) according to the manufacturer’s instructions.
Dual-luciferase assays of transformed tobacco leaves
The full-length cDNAs of TaERF3 and TaERF115 were ampli- fied using the primers TaERF3-1300-F/R and TaERF115-1300-F/R
![]()
(Supplementary Table S2) and separately cloned into the effector vector pCAMBIA1300 under the control of CaMV 35S. The TaSDIR1-4A promoter fragment was amplified from H10 and L14 using the primers Hap-LUC-F/R (Supplementary Table S2) and separately ligated into the reporter vector pGreen II 0800-LUC. The effector and reporter con- structs were transformed into GV3101 cells using pSoup, and transformed into leaves of 4-week-old Nicotiana tabacum plants by co-infiltration.The activities of firefly luciferase (LUC) and Renilla luciferase (REN) were measured using the Dual-Glo® Luciferase Assay System (E2920, Promega) with a multimode reader (TriStar2 S LB942) at 48 h after infiltration. The promoter activity was calculated as the ratio of LUC to REN, and the ratio in leaves transformed with the empty vector (pCAMBIA1300/ pGreen II 0800-LUC) was set to 1.
Results
TaSDIR1-4A is a functional E3 ligase
Two copies of TaSDIR1 were isolated from the chromosome group 4 of subgenomes A and D in wheat, and hence were named as TaSDIR1-4A and TaSDIR1-4D. TaSDIR1-4D is known to be involved in responses to abiotic stress (Wang et al., 2018). The cDNA sequence of TaSDIR1-4A obtained from cv. H10 consisted of eight exons and seven introns and encoded a protein containing 280 amino acids.TaSDIR1-4A, belonging to the RING finger domain family, contained two transmembrane domains in the N-terminal region, and a C3H2C3-type RING domain in the C-terminal region (Supplementary Fig. S1).
Previous studies have shown that the RING finger- containing protein AtSDIR1 functions as an E3 ligase (Zhang et al., 2007). An in vitro E3 ubiquitin ligase activity assay was therefore performed to test whether TaSDIR1-4A had E3 ligase activity, using AtSDIR1 as a positive control. Ubiquitination activity was observed in the presence of 6×His tag ubiquitin (Ub), E1 (from wheat), E2 (UBCh5b) and purified MBP- AtSDIR1 or MBP-TaSDIR1-4A proteins using an anti-Ub antibody (Fig. 1). An anti-MBP blot analysis also indicated that MBP-AtSDIR1 and MBP-TaSDIR1-4A were ubiquitinated. However, with MBP alone, or in the absence of E1, E2, E3, or Ub, no polyubiquitination was detected. The highly conserved RING finger domain of SDIR1, from Cys211 to Cys241 in Arabidopsis, has E3 ubiquitin ligase activity.The equivalent func- tional region in TaSDIR1-4A was from Cys221 to Cys251.The RING motif was essential for the E3 ligase activity of AtSDIR1. Our previous work had generated an allele with a single amino acid substitution by mutagenizing His-234 to Tyr (H234Y) in AtSDIR1, which abolished the E3 ligase activity (Zhang et al., 2007). Presumably the mutation disrupted the RING domain. A similar mutation in TaSDIR1-4A, His-244-Tyr, was produced, and again an in vitro ubiquitination assay indicated that the E3 ligase activity was completely disrupted (Fig. 1). These results indicated that TaSDIR1-4A has E3 ligase activity and that an intact RING domain is required for enzyme activity.
Sequence polymorphism assays, genetic mapping, and association analysis
Sequence polymorphism assays indicated that there was no nucleotide polymorphism in the TaSDIR1-4A coding re- gion, but two nucleotide variation sites were detected in the
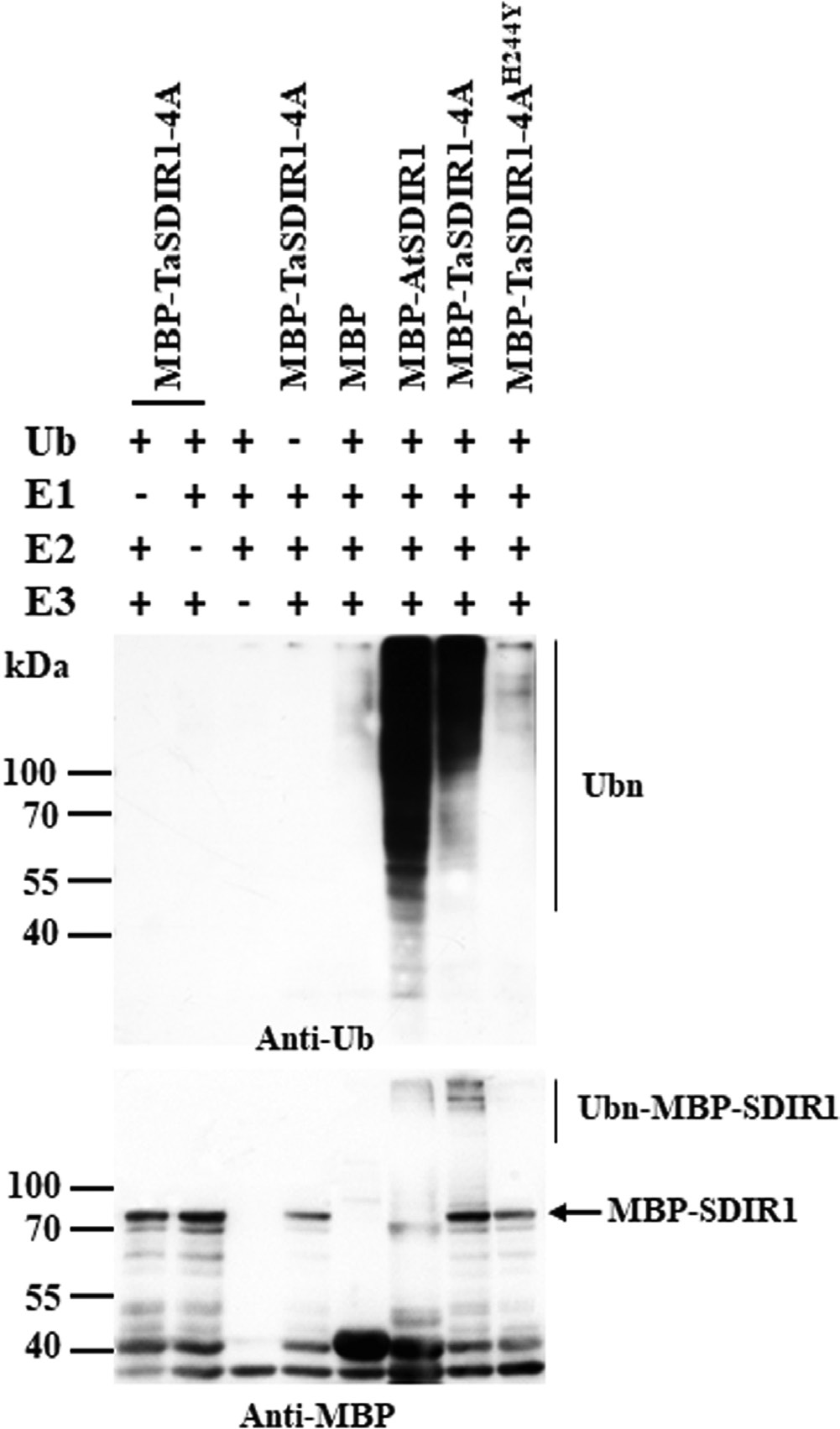
![]() Fig. 1. E3 ubiquitin ligase activity of wheat TaSDIR1-4A. The fusion proteins MBP-TaSDIR1-4A and its mutant form MBP-TaSDIR1-4AH244Y were assayed for E3 activity in the presence of E1 (from wheat), E2 (UBCh5b), and 6×His tag ubiquitin (Ub). The molecular masses of the marker proteins are shown (kDa). MBP was used as a negative control and MBP-AtSDIR1 was used as a positive control. Samples were resolved by 10% SDS-PAGE. An anti-Ub antibody was used to detect His tag ubiquitin (top image), and the anti-MBP antibody was used for detection of maltose fusion proteins (bottom image).
Fig. 1. E3 ubiquitin ligase activity of wheat TaSDIR1-4A. The fusion proteins MBP-TaSDIR1-4A and its mutant form MBP-TaSDIR1-4AH244Y were assayed for E3 activity in the presence of E1 (from wheat), E2 (UBCh5b), and 6×His tag ubiquitin (Ub). The molecular masses of the marker proteins are shown (kDa). MBP was used as a negative control and MBP-AtSDIR1 was used as a positive control. Samples were resolved by 10% SDS-PAGE. An anti-Ub antibody was used to detect His tag ubiquitin (top image), and the anti-MBP antibody was used for detection of maltose fusion proteins (bottom image).
TaSDIR1-4A promoter region, a 2-bp InDel at –412 to –411 and a SNP at –395(Supplementary Fig. S2). Two haplotypes, Hap-4A-1 and Hap-4A-2, were identified (Fig. 2A, B) and a functional marker (FM) based on SNP-395 (G/A) was able to distinguish between them (Fig. 2C). The marker contained a mismatch in the downstream primer that produced a recogni- tion site for the restriction enzyme Age I.
Using diploid and tetraploid wheat relatives and a set of Chinese Spring nullisomic-tetrasomic lines, TaSDIR1-4A was found to be located on chromosome 4A (Supplementary Fig. S3A), and the functional marker was used to genotype the doubled-haploid (DH) lines in order to further map its position. Linkage analysis located a QTL for TGW between this functional marker and AX-111611657 (Supplementary Fig. S3B). Using the same DH lines, a QTL for TGW and the
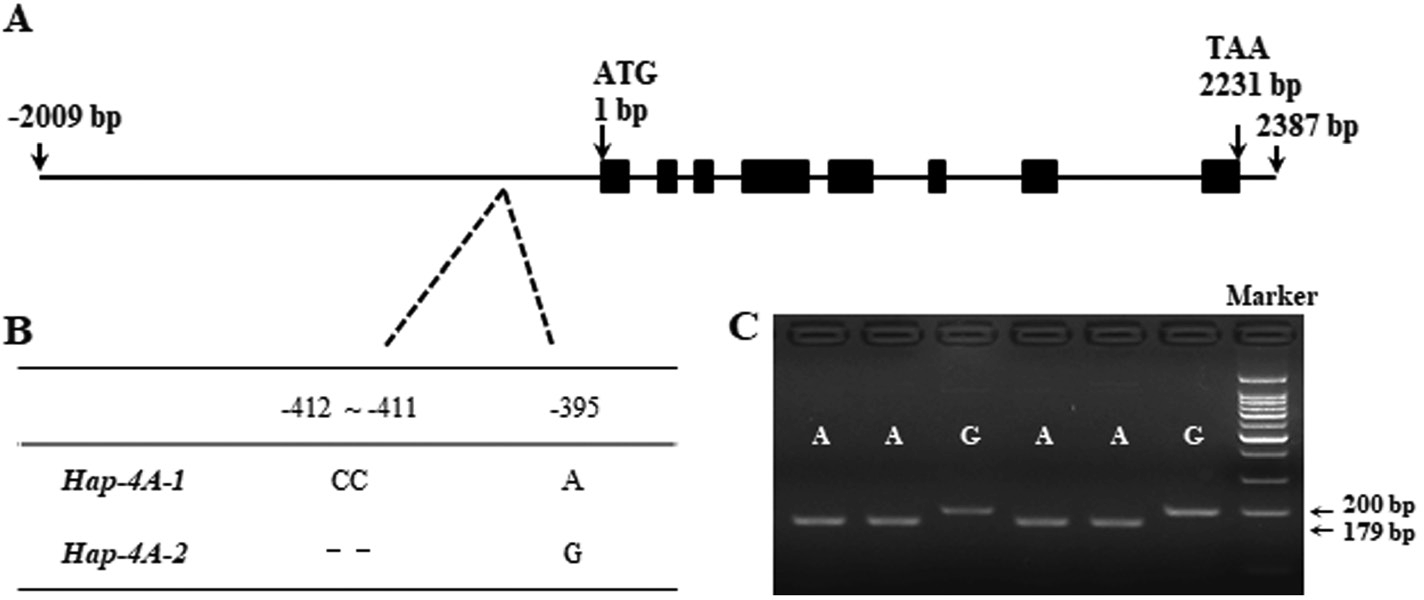
![]() Fig. 2. Nucleotide polymorphisms and development of a functional marker in wheat TaSDIR1-4A. (A) Schematic diagram of the TaSDIR1-4A structure. The ATG start codon was designated as position 1 bp. (B) Polymorphic sites were detected in the promoter regions of TaSDIR1-4A. (C) A dCAPS marker was developed based on the –395 bp single-nucleotide polymorphism (G/A). Digestion of the amplified 200-bp fragment with Age I produced fragments of 179 bp and 21 bp for accessions with the haplotype Hap-4A-1 (A), whereas this fragment was not digested in accessions with the haplotype
Fig. 2. Nucleotide polymorphisms and development of a functional marker in wheat TaSDIR1-4A. (A) Schematic diagram of the TaSDIR1-4A structure. The ATG start codon was designated as position 1 bp. (B) Polymorphic sites were detected in the promoter regions of TaSDIR1-4A. (C) A dCAPS marker was developed based on the –395 bp single-nucleotide polymorphism (G/A). Digestion of the amplified 200-bp fragment with Age I produced fragments of 179 bp and 21 bp for accessions with the haplotype Hap-4A-1 (A), whereas this fragment was not digested in accessions with the haplotype
Hap-4A-2 (G). Marker, 100-bp DNA ladder.
Table 1. TaSDIR1-4A haplotypes associated with agronomic traits across 10 environments
![]()
TaSDIR1-4A is a negative regulator of grain size
To further examine the role of TaSDIR1-4A in wheat we si-
Year planted
Site Conditions Effect on 1000-grain
weight
![]()
![]() P-value PVE (%)
P-value PVE (%)
lenced its transcription by BSMV-mediated VIGS in the cul-
tivar Chinese Spring. BSMV-associated chlorotic stripe mosaic symptoms appeared in newly emerged leaves 10 d post in-
2010 CP DS 3.56×10–6*** 10.78
CP WW 8.38×10–6*** 8.59
SY DS 4.09×10–5*** 8.20
SY WW 1.74×10–4*** 6.04
2011 SY DS 0.0264* 4.41
SY WW 0.0013*** 4.11
2012 CP DS 4.57×10–6*** 9.48
|
CP |
WW |
0.03* |
4.03 |
SY |
DS |
7.48×10–6*** |
8.47 |
|
|
SY |
WW |
3.03×10–5*** |
8.91 |
PVE, phenotypic variation explained. Sites: CP, Changping; SY, Shunyi. Conditions: DS, drought-stressed; WW, well-watered. Significant differences between the TaSDIR1-4A haplotypes were determined using Student’s t-test: *P<0.05, ***P<0.001.
TGW-related trait stem water-soluble carbohydrates had pre- viously been identified in an adjacent interval flanked by the markers Xgwm601 and WMC420 (Yang et al., 2007; Li et al., 2012a).The marker Xgwm601 was 3.56 cM from TaSDIR1-4A. Another QTL for the TGW-related trait chlorophyll content is also present in this region (Li et al., 2010a). This indicated that TaSDIR1-4A might have a connection with grain weight. Population 2 was used to examine the associations be- tween the TaSDIR1-4A haplotypes and agronomic traits. Results of sequence polymorphism analysis of 262 acces- sions (Supplementary Table S4) indicated that variation at the TaSDIR1-4A locus was significantly associated with TGW in all the 10 environments that were examined (Table 1). Hap-4A-1 and Hap-4A-2 accounted for 36.0% and 64.0% of entries in this population, respectively. Accessions with Hap-4A-2 had a higher mean TGW than accessions with Hap-4A-1 (Fig. 3A), and thus it represented a more favorable haplotype. These differences were also found in
Populations 1 and 4 (Fig. 3B, C).
oculation with the virus (Supplementary Fig. S4A). Flag leaves were harvested at anthesis and RT-PCR showed a 60% de- cline in transcripts in the BSMV:TaSDIR1-4A (silenced) lines compared with the BSMV:GFP lines (Fig. 4A). Quantitative real-time PCR was performed to confirm the specificity of knock-down of the expression of TaSDIR1-4A, no significant difference was observed in the expression of TaSDIR1-4D (the gene with the highest sequence similarity to TaSDIR1-4A) between the control and BSMV:TaSDIR1-4A lines (Supplementary Fig. S4B). Measurements after harvest showed that the silenced lines had larger grain size and higher TGW than the BSMV:GFP lines (Fig. 4B–D) whilst the BSMV-free lines had larger grain size and higher TGW than the BSMV- infected lines. Assuming that damage from BSMV infection would lead to smaller grain size and lower TGW, these results indicated that TaSDIR1-4A had a negative role in the deter- mination of TGW.
To further confirm its effect on grain size and TGW,
TaSDIR1-4A was overexpressed in the wild-type and the sdir1-1 mutant of Arabidopsis and relative expression was de- termined by semi-quantitative PCR (Supplementary Fig. S5). The sdir1-1 line had larger seeds whilst the TaSDIR1-4A- overexpression lines in both the wild-type and mutant back- ground had smaller seeds than the non-transgenic controls (Fig. 4E).
TaSDIR1-4A expression patterns
Because sequence polymorphism sites were located in the TaSDIR1-4A promoter region, we detected effects of vari- ation on expression. We first performed real-time PCR on a range of tissues to determine expression patterns in wheat at the flowering stage, and found that TaSDIR1-4A was
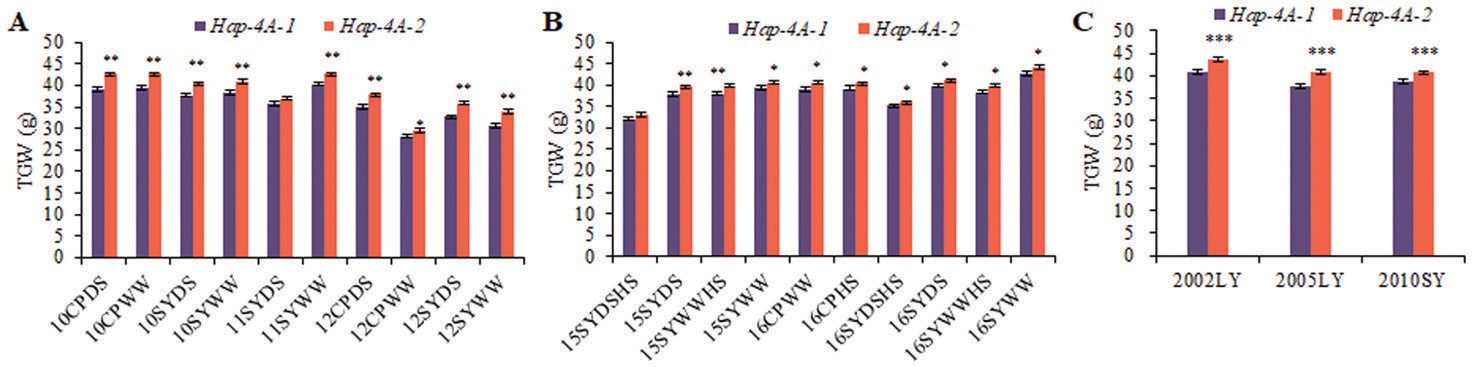
![]() Fig. 3. Comparisons of 1000-grain weight (TGW) in the two wheat TaSDIR1-4A haplotypes Hap-4A-1 and Hap-4A-2. (A) Comparisons in Population 2 (262 accessions) grown in 10 different environments: either at Changping (CP) or Shunyi (SY), and under drought-stressed (DS) or well-watered (WW) conditions, planted in 2010, 2011, or 2012. (B) Comparisons in Population 1 (doubled-haploid) grown in 10 different environments: either at CP or SY, and under DS, WW, or heat-stressed (HS) conditions, planted in 2015 or 2016. (C) Comparisons in Population 4 (348 modern cultivars) grown in three different environments: SY planted in 2010 or Luoyang (LY) planted in 2002 or 2005. Data are means (±SE), n=3. Significant differences between the haplotypes were determined using Student’s t-test: *P<0.05, **P<0.01, and ***P<0.001.
Fig. 3. Comparisons of 1000-grain weight (TGW) in the two wheat TaSDIR1-4A haplotypes Hap-4A-1 and Hap-4A-2. (A) Comparisons in Population 2 (262 accessions) grown in 10 different environments: either at Changping (CP) or Shunyi (SY), and under drought-stressed (DS) or well-watered (WW) conditions, planted in 2010, 2011, or 2012. (B) Comparisons in Population 1 (doubled-haploid) grown in 10 different environments: either at CP or SY, and under DS, WW, or heat-stressed (HS) conditions, planted in 2015 or 2016. (C) Comparisons in Population 4 (348 modern cultivars) grown in three different environments: SY planted in 2010 or Luoyang (LY) planted in 2002 or 2005. Data are means (±SE), n=3. Significant differences between the haplotypes were determined using Student’s t-test: *P<0.05, **P<0.01, and ***P<0.001.
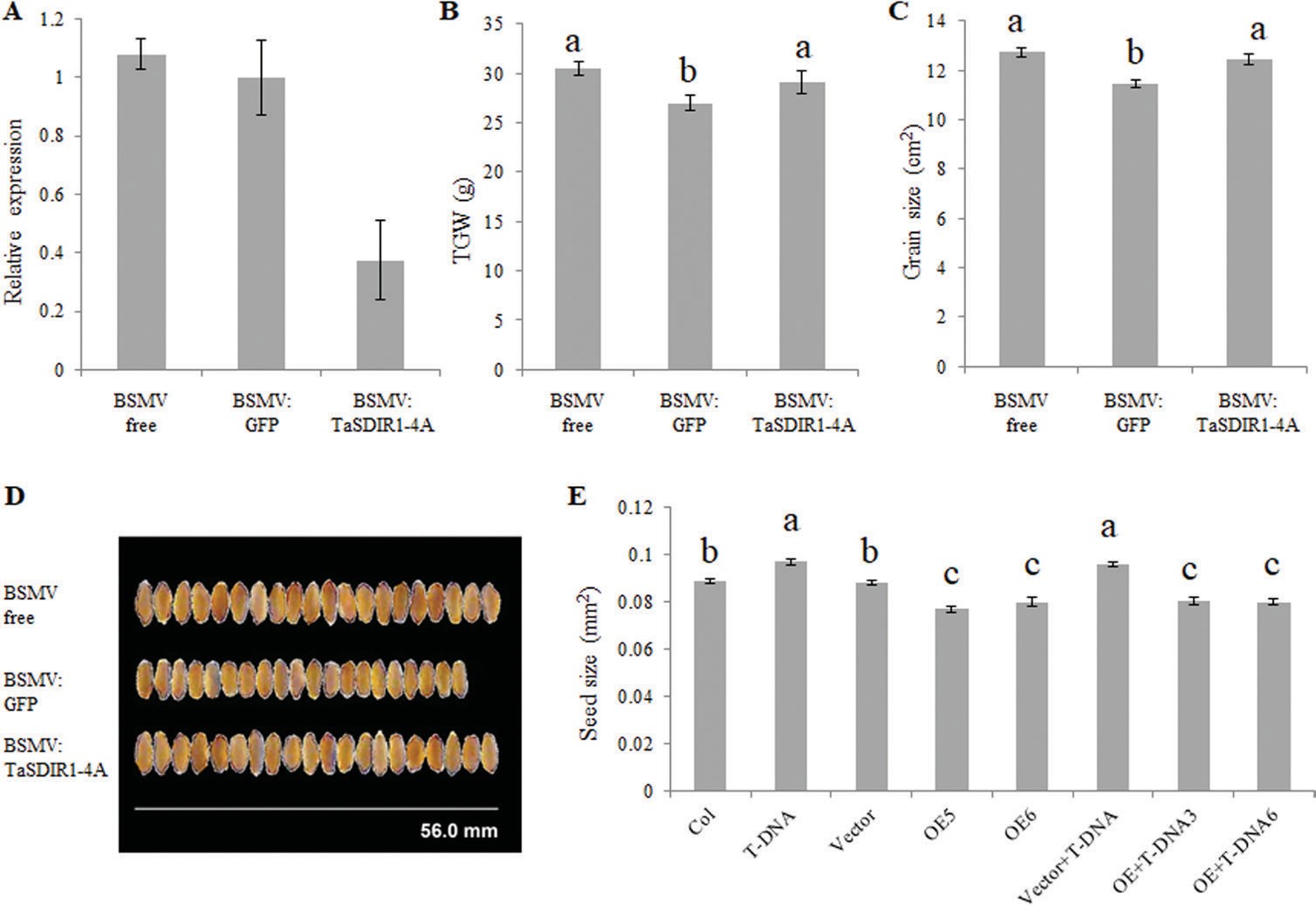
Fig. 4. Phenotypic analysis of TaSDIR1-4A-silenced wheat and transgenic Arabidopsis. Virus-induced gene silencing (VIGS) of wheat was undertaken using a series of recombinant barley stripe mosaic virus (BSMV) vectors. (A) Relative expression of TaSDIR1-4A showing that transcription was lower in BSMV:TaSDIR1-4A plants than BSMV-free and BSMV:GFP plants. (B) Thousand-grain weight (TGW), (C) grain size, and (D) representative images of the grains. (E) Seed size in Arabidopsis plants in the Col-0 wild-type and T-DNA insertion mutant sdir1-1 backgrounds overexpressing (OE) TaSDIR1-4A. Vector, empty vector control. Significant differences between means were determined using one-way ANOVA followed by Tukey’s test (P<0.05).
constitutively expressed in all tissues, with highest expression levels in the flag leaves (Fig. 5A). We then randomly selected 12 accessions of each of the two haplotypes from Populations 1 and 2 and planted them in the field (Supplementary Table S3). Flag leaves were collected at anthesis and the expression levels of TaSDIR1-4A were determined. The mean relative expression level of the Hap-4A-1 accessions was 2.21-fold
greater than that of the Hap-4A-2 accessions in Population 1 (Fig. 5B), and 2.77-fold greater in Population 2 (Fig. 5C). In agreement with the results of the association analysis, accessions with Hap-4A-1 had higher gene expression and lower TGW relative to those with Hap-4A-2, indicating that lower expres- sion of TaSDIR1-4A Hap-4A-2 resulted in higher TGW, and that TaSDIR1-4A plays a negative role in regulating TGW.
![]()
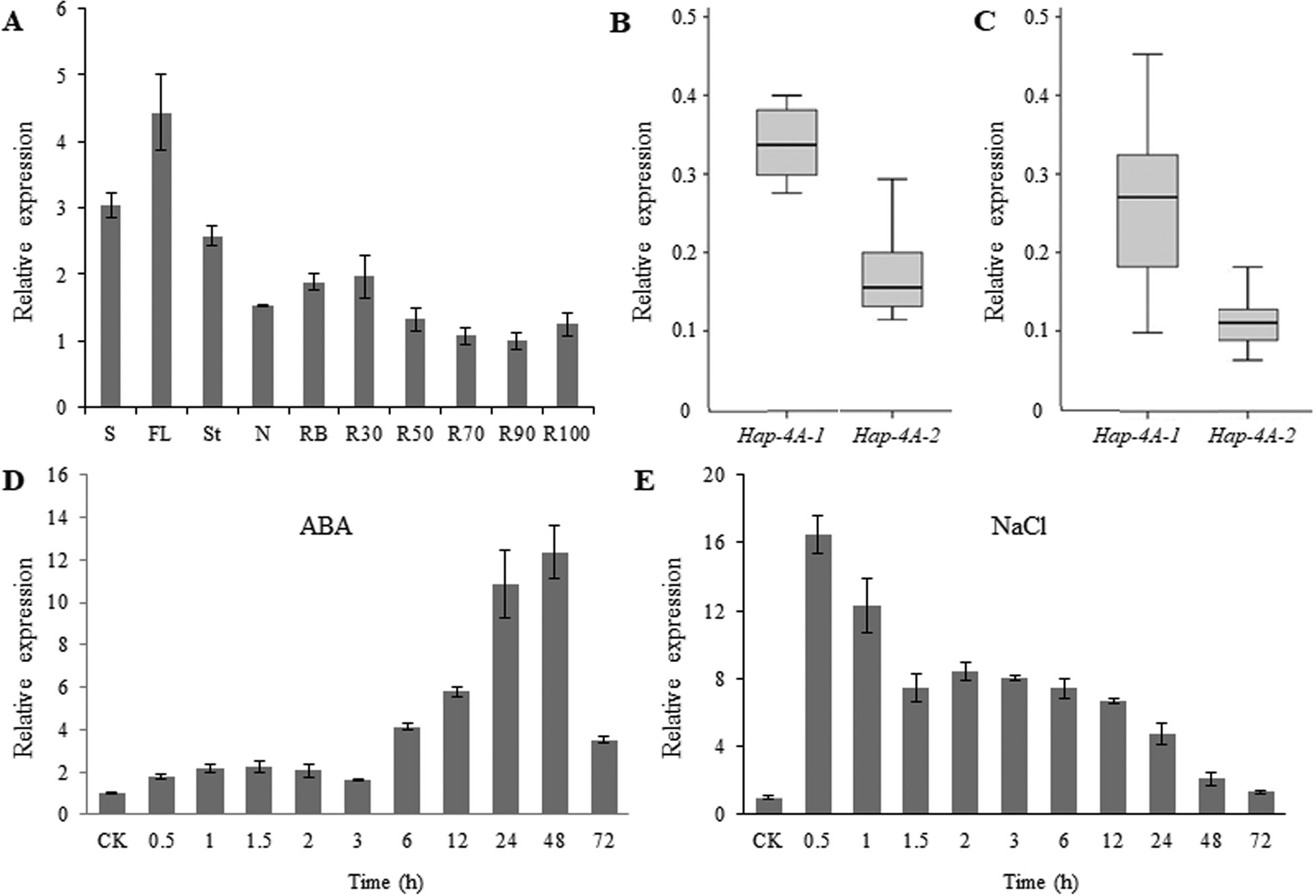
![]() Fig. 5. Expression patterns of the wheat TaSDIR1-4A haplotypes Hap-4A-1 and Hap-4A-2. (A) Expression patterns of TaSDIR1-4A in different tissues of cv. H10 plants as measured by real-time PCR. S, spikes at flowering; FL, flag leaves; St, stems; N, nodes; RB, root bases; R30, roots from 0–30 cm depth; R50, roots from 30–50 cm; R70, roots from 50– cm; R90, roots from 70–90 cm; R100, roots from 90–100 cm. The values are relative to R90, which was set as 1. (B, C) Expression of Hap-4A-1 and Hap-4A-2 in (B) Population 1 and (C) Population 2. For each haplotype 12 accessions were
Fig. 5. Expression patterns of the wheat TaSDIR1-4A haplotypes Hap-4A-1 and Hap-4A-2. (A) Expression patterns of TaSDIR1-4A in different tissues of cv. H10 plants as measured by real-time PCR. S, spikes at flowering; FL, flag leaves; St, stems; N, nodes; RB, root bases; R30, roots from 0–30 cm depth; R50, roots from 30–50 cm; R70, roots from 50– cm; R90, roots from 70–90 cm; R100, roots from 90–100 cm. The values are relative to R90, which was set as 1. (B, C) Expression of Hap-4A-1 and Hap-4A-2 in (B) Population 1 and (C) Population 2. For each haplotype 12 accessions were
randomly selected for measurement. (D) Expression patterns of TaSDIR1-4A following ABA treatment. Seedlings of cv. H10 at 2 weeks old were sprayed with 50 μM ABA solution and were sampled at 0 h (control, CK) and at 0.5–72 h. Expression is relative to the value in the control, which was set as 1. (E) Expression patterns of TaSDIR1-4A following NaCl treatment. Seedlings of cv. H10 at 2 weeks old were treated with 250 mM NaCl and were sampled at 0 h (CK) and at 0.5–72 h. Expression is relative to the value in the control, which was set as 1. In all cases GAPDH was used as the internal control. All data are means (±SE) of three biological replicates.
To determine whether TaSDIR1-4A plays a role in balancing the salt-stress response and yield, we examined re- sponses to treatments with salt and ABA using real-time PCR. TaSDIR1-4A expression was induced by ABA (Fig. 5D) and transcription levels were at their highest (12-fold greater than the control) after 48 h of treatment. TaSDIR1-4A expression was also induced by salt and reached its highest level (16-fold greater than the control) after 0.5 h of treatment, following which it declined (Fig. 5E). These results demonstrated that TaSDIR1-4A was up-regulated in response to ABA and salt stress.
TaERF3 regulates TaSDIR1-4A expression by binding to the promoter region
To further investigate the effects of the two polymorphic sites in the promoter region of TaSDIR1-4A, we first identified differences in cis-element between the two haplotypes. Using the Plant Transcription Factor Database (http://planttfdb.cbi. pku.edu.cn/), an ERF-binding site (CCCCGCCG) was iden- tified at 395 bp upstream of the ATG translation start site in
Hap-4A-2; the binding site was absent in Hap-4A-1 due an A-to-G difference (CCCCACCG).
Yeast one-hybrid assays were performed to determine whether TaERF would bind to the promoter region of TaSDIR1-4A by cloning the TaERF DNA binding region into the pB42AD vector. Each of the six fragments of the TaSDIR1-4A promoter region was cloned into pLacZi (Fig. 6A) and their interactions with TaERF-BD were tested by transforming it into yeast strain EGY48 cultured on selective media with x-α-gal and lacking Ura and Leu. In the presence of the TaSDIR1-4A promoter region from cv. L14 or the 3×GCC Box (either from cv. H10 or L14), TaERF-BD bound to and activated the LacZ reporter gene, whereas in the presence of the TaSDIR1-4A promoter re- gion from H10 or the 3×ACC Box (neither from H10 or L14) it did not (Fig. 6A, B). These results suggested that the TaERF-BD could bind to the promoter of Hap-4A-2 but not to that of Hap-4A-1 due to the GCC/ACC Box differ- ence rather than to the 2-bp InDel difference. These differ- ences in binding might be responsible for the differences in the expression levels of two haplotypes.
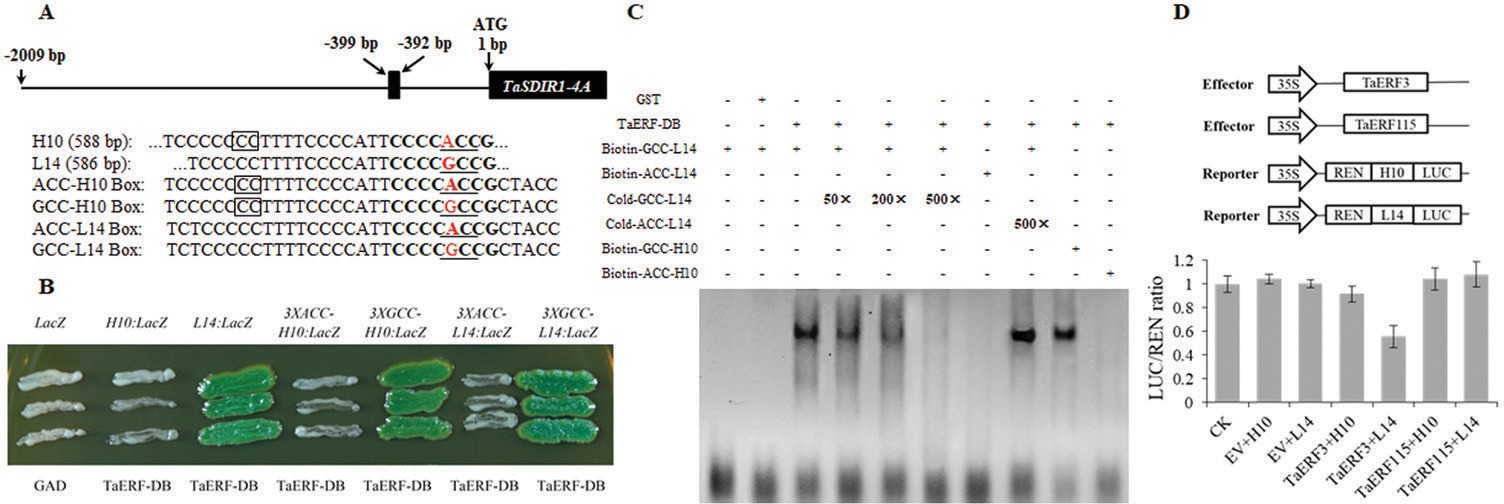
![]() Fig. 6. TaERF3 binds to the wheat TaSDIR1-4A Hap-4A-2 promoter region. (A) Schematic diagram of the GCC Box (–399 to –392 bp) in the Hap-4A-2 promoter. Letters in bold indicate the GCC Box regions, underlined letters indicate the GCC Box, letters in red are the single-nucleotide polymorphism sites (G/A), and the boxes indicate the 2-bp InDel sites. The fragments from the TaSDIR1-4A promoter regions of the cultivars H10 and L14 are shown, together with the 32-bp ACC Box from H10 (ACC-H10 Box), the same fragment but with A-to-G mutation (GCC-H10 Box), the 32-bp GCC Box from L14 (GCC-L14 Box), and the same fragment but with G-to-A mutation (ACC-L14 Box). (B) GAD-TaERF-BD activated expression in yeast of the LacZ reporter gene driven by the Hap-4A-2 (L14) promoter and the GCC Box (either from H10 or L14), rather than the Hap-4A-1 (H10) promoter and the 3×ACC Box (either from H10 or L14). (C) EMSA of the purified TaERF-BD protein and the GCC Box probe. Lane 1 contains biotin-labelled GCC-L14 Box probe, Lane 2 contains purified GST and biotin-labelled GCC-L14 Box probe, Lane 3 contains the TaERF-BD protein and biotin-labelled GCC-L14 Box probe, Lane 4 contains the TaERF-BD protein, biotin-labelled GCC-L14 Box probe, and 50× unlabeled (cold) GCC-L14 Box probe, Lane 5 contains the TaERF-BD protein, biotin-labelled GCC-L14 Box probe, and 200× cold GCC-L14 Box probe, Lane 6 contains the TaERF-BD protein, biotin-labelled GCC-L14 Box probe, and 500× cold GCC-L14 Box probe, Lane 7 contains the TaERF-BD protein, biotin-labelled ACC-L14 Box probe, Lane 8 contains the TaERF-BD protein, biotin-labelled GCC-L14 Box probe, and 500× unlabeled ACC-L14 Box probe, Lane 9 contains the TaERF-BD protein and
Fig. 6. TaERF3 binds to the wheat TaSDIR1-4A Hap-4A-2 promoter region. (A) Schematic diagram of the GCC Box (–399 to –392 bp) in the Hap-4A-2 promoter. Letters in bold indicate the GCC Box regions, underlined letters indicate the GCC Box, letters in red are the single-nucleotide polymorphism sites (G/A), and the boxes indicate the 2-bp InDel sites. The fragments from the TaSDIR1-4A promoter regions of the cultivars H10 and L14 are shown, together with the 32-bp ACC Box from H10 (ACC-H10 Box), the same fragment but with A-to-G mutation (GCC-H10 Box), the 32-bp GCC Box from L14 (GCC-L14 Box), and the same fragment but with G-to-A mutation (ACC-L14 Box). (B) GAD-TaERF-BD activated expression in yeast of the LacZ reporter gene driven by the Hap-4A-2 (L14) promoter and the GCC Box (either from H10 or L14), rather than the Hap-4A-1 (H10) promoter and the 3×ACC Box (either from H10 or L14). (C) EMSA of the purified TaERF-BD protein and the GCC Box probe. Lane 1 contains biotin-labelled GCC-L14 Box probe, Lane 2 contains purified GST and biotin-labelled GCC-L14 Box probe, Lane 3 contains the TaERF-BD protein and biotin-labelled GCC-L14 Box probe, Lane 4 contains the TaERF-BD protein, biotin-labelled GCC-L14 Box probe, and 50× unlabeled (cold) GCC-L14 Box probe, Lane 5 contains the TaERF-BD protein, biotin-labelled GCC-L14 Box probe, and 200× cold GCC-L14 Box probe, Lane 6 contains the TaERF-BD protein, biotin-labelled GCC-L14 Box probe, and 500× cold GCC-L14 Box probe, Lane 7 contains the TaERF-BD protein, biotin-labelled ACC-L14 Box probe, Lane 8 contains the TaERF-BD protein, biotin-labelled GCC-L14 Box probe, and 500× unlabeled ACC-L14 Box probe, Lane 9 contains the TaERF-BD protein and
biotin-labelled GCC-H10 Box probe, and Lane 10 contains the TaERF-BD protein and biotin-labelled ACC-H10 Box probe. The upper bands show that the TaERF-BD protein binds to the biotin-labelled GCC Box probe and the lower bands show the free probes. (D) Dual-luciferase assay of transformed tobacco leaves to examine the interaction between TaERFs and the TaSDIR1-4A promoter. Agrobacterium transformed with the vector pSoup harboring reporter and effector constructs, or with the empty vector (EV), were co-infiltrated into the leaves. Schematic diagrams of the effector and reporter constructs are shown. TaERF3 or TaERF115 was cloned into the effector construct pCAMBIA1300 and the promoter fragments from H10 or L14 were inserted into the reporter vector pGreen II 0800-LUC. Promoter activities are shown as the ratio of LUC to REN, and are relative to the control (CK) value, which was set to 1. Data are means (±SE) of three biological replicates.
![]()
Following induction and purification of theTaERF-BD protein, EMSAs were carried out to verify that it binds to the TaSDIR1-4A promoter region (Supplementary Fig. S6). Compared with the free probe, a slower-migrating DNA-binding band was detected with the addition of TaERF-BD and the biotin-labelled GCC Box probe (from either H10 or L14) (Fig. 6C), whereas there was no band in the GST control. The biotin-labelled DNA-binding band was diminished as the concentration of the unlabeled GCC Box probe was increased.The TaERF-BD could not bind to the biotin-labelled ACC Box probe from either H10 nor L14 and the unlabeled ACC Box probe did not compete with the labelled GCC Box probe. These results confirmed that the TaERF-BD could bind to the GCC Box region in the Hap-4A-2 promoter but not to the ACC Box region in the Hap-4A-1 promoter, and that this was due to the GCC/ACC Box difference rather than to the 2-bp InDel difference.
To examine which TaERF affects the expression of
TaSDIR1-4A, we selected TaERF3, an ortholog of OsERF3, as the candidate protein based on previous reports that OsERF3 has EAR motif-mediated transcriptional repression activity and regulates yield-related genes (Ohta et al., 2001; Kagale et al., 2010; Wang et al., 2011).TaERF3 is known to have relatively high ex- pression levels in flower organs (Supplementary Fig. S7). It has been reported that OsERF115 can bind to the GCC box while not possessing transcriptional activation or repression activity, and OsERF115 also shows higher expression levels during the grain
development period and is associated with yield (Xu et al., 2016). Hence TaERF115, an ortholog of OsERF115, was selected as the negative control. Dual luciferase assays showed that LUC activity was lower in the presence of both the TaERF3 effector and the TaSDIR1-4A promoter from L14 reporter constructs than in the negative control (Fig. 6D). In contrast, LUC activity was similar to the negative control in the presence of both the TaERF3 effector and the TaSDIR1-4A promoter from H10 reporter constructs, or in the presence of the TaERF115 effector and the TaSDIR1-4A promoter from H10/L14 reporter constructs. These results sug- gested that TaERF3 only acts as a transcriptional repressor of TaSDIR1-4A from L14 and not of TaSDIR1-4A from H10.
Geographic and temporal distribution of TaSDIR1-4A haplotypes across different wheat production zones in China
Genotypes of accessions in Population 3 (157 landraces) and Population 4 (348 modern cultivars) covering 10 different wheat production zones across the whole of China were used to investigate the geographic and temporal distributions of the TaSDIR1 haplotypes. Hap-4A-1 was the dominant haplo- type in landraces from all 10 zones, especially Zones V and IX where all the accessions were of this haplotype (Fig. 7). In con- trast, in modern cultivars the frequency of Hap-4A-2 was gen- erally higher, except in Zones V and IX, where the frequencies
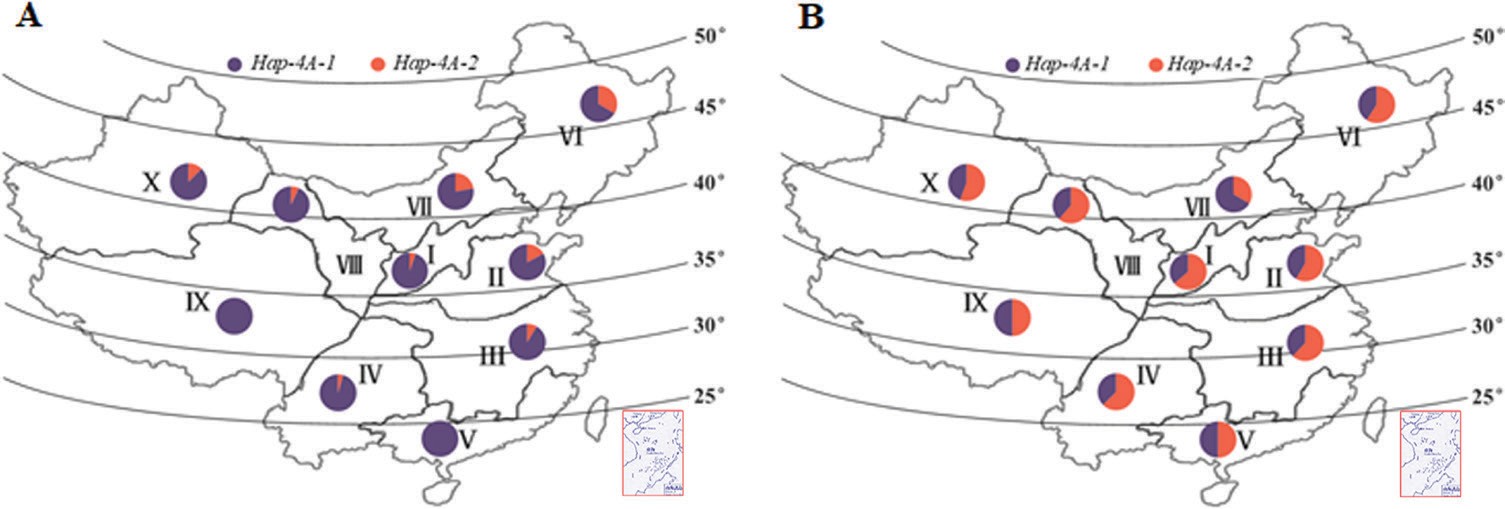
![]() Fig. 7. Distribution of the TaSDIR1-4A haplotypes Hap-4A-1 and Hap-4A-2 in 10 wheat-producing regions across China. (A) Proportions of the two haplotypes in 157 Chinese landraces and (B) in 348 modern cultivars. Winter wheat regions: I, Northern; II, Yellow and Huai River valleys; III, Low and middle Yangtze River valley; IV, Southwestern; V, Southern. Spring wheat regions: VI, Northeastern; VII, Northern; VIII, Northwestern. Spring–winter wheat regions: IX, Qinghai–Tibet; X, Xinjiang.
Fig. 7. Distribution of the TaSDIR1-4A haplotypes Hap-4A-1 and Hap-4A-2 in 10 wheat-producing regions across China. (A) Proportions of the two haplotypes in 157 Chinese landraces and (B) in 348 modern cultivars. Winter wheat regions: I, Northern; II, Yellow and Huai River valleys; III, Low and middle Yangtze River valley; IV, Southwestern; V, Southern. Spring wheat regions: VI, Northeastern; VII, Northern; VIII, Northwestern. Spring–winter wheat regions: IX, Qinghai–Tibet; X, Xinjiang.

Fig. 8. Frequencies of the wheat TaSDIR1-4A haplotypes Hap-4A-1
and Hap-4A-2 in Population 4 (348 modern cultivars) together with their corresponding 1000-grain weights plotted against the decades in which the cultivars were released. The data for TGW are from Hao et al. (2011), n=3.
of the two haplotypes were approximately equal, and in Zone VII, the Northern Spring Wheat Zone, where the frequency of Hap-4A-2 was lower than that of Hap-4A-1.
The meanTGW in modern wheat varieties released in China from the pre-1950s to the 1990s has increased from 32.98 g to
42.97 g, and this has coincided with the proportion of haplo- type Hap-4A-2 increasing from 25.00% to 78.12% whilst that of Hap-4A-1 declined from 75.00 to 21.88% (Fig. 8). This in- dicates that Hap-4A-2 was positively selected in Chinese wheat breeding programs.
Discussion
TaSDIR1-4A is a functional RING finger E3 ligase with an unknown substrate
We have shown that TaSDIR1-4A is a functional RING finger E3 ligase (Fig. 1). Previous studies have demonstrated that AtSDIR1 targets SDIR1-INTERACTING PROTEIN1 for
degradation in modulation of the response to salt stress and in ABA signaling (Zhang et al., 2015). The rice RING-type protein GW2, which has E3 ubiquitin ligase activity, acts as a negative regulator of grain width, weight, and yield (Song et al., 2007). TaGW2 also negatively regulates grain width and weight (Hong et al., 2014; Jaiswal et al., 2015; Simmonds et al., 2016; Liu et al., 2020). However, the substrate on which TaSDIR1-4A acts in its control of grain size, and any rela- tionship between it and the TaGW2 substrate, requires further study.
TaSDIR1-4A is a negative regulator of grain size
Previous studies have demonstrated that SDIR1 is induced by stress and has a positive role in abiotic stress responses, but the role of SDIR1 in grain size development has been un- known (Zhang et al., 2008; Liu et al., 2013; Oh et al., 2017). In this study, we have clearly shown that TaSDIR1-4A is a negative regulator of grain size and weight in two ways. Firstly, silencing of TaSDIR1-4A led to increased TGW in wheat and overexpression of TaSDIR1-4A in Arabidopsis resulted in reduced seed size (Fig. 4). Secondly, wheat acces- sions with the haplotype Hap-4A-2 had lower TaSDIR1-4A expression and higher TGW than accessions with Hap-4A-1 (Figs 3, 5B, C).
TaSDIR1-4A might balance the relationship between stress responses and grain yield
Given that TaSDIR1-4A was up-regulated in response to ABA and salt-stress treatments (Fig. 5D, E) and that TaSDIR1-4A was a negative regulator of grain size (Figs 3, 4, 5B, C), we propose that TaSDIR1-4A may play a role in balancing responses to stress and yield. Under normal conditions, lower expression levels of TaSDIR1-4A may contribute to higher yield in wheat, whilst under abiotic stress conditions up-regulation of TaSDIR1-4A may help to overcome the stress but at the cost of reduced grain yield.
![]()
This suggests that TaSDIR1-4A is the central control unit of complex networks that balance responses under constantly changing environmental conditions.
TaERF3 affects the expression of TaSDIR1-4A
haplotypes
Ethylene response factor (ERF) specifically binds to a GCC Box and has important functions in the control of grain size and yield (Ohto et al., 2005; Aya et al., 2014; Müller and Munné- Bosch, 2015). For example, OsERF3 can bind to the promoter of LRK6, a rice yield-related gene, which might lead to its dif- ferential expression (Kagale et al., 2010;Wang et al., 2011). Salt- responsive ERF1 (SERF1) is a negative regulator of grain size in rice (Schmidt et al., 2014). And the closely related rice ERF genes AP37 and AP59 have different effects on yield (Oh et al., 2009). Tobacco NtERF3 and Arabidopsis AtERF3, which has an ERF-associated amphiphilic repression (EAR) motif, are active repressors of transcription (Ohta et al., 2001). Our results demonstrated that TaERF3, which also has EAR motif, could bind to the promoter region of the TaSDIR1-4A haplotype Hap-4A-2 and repress its expression, but that it did not do so for Hap-4A-1 (Fig. 6). Accessions possessing Hap-4A-2 had lower TaSDIR1-4A expression levels and greater TGWs com- pared to those possessing Hap-4A-1. These findings suggest that TaERF3 is a positive regulator for TGW (Supplementary Fig. S8), but its biological functioning needs further study.
A stable and effective molecular marker for breeding
Pyramiding elite alleles for important agronomic traits through marker-assisted selection is an effective approach to accelerate crop breeding. In this study, we designed primers specific to the A genome for detection of the two TaSDIR1-4A haplo- types. The dCAPS marker that we identified was based on a single-nucleotide polymorphism and it distinguished between the haplotypes associated with the differences in TGW. Hence, it can be used in future marker-assisted breeding for greater TGW, although we recognize that the frequency of the favor- able Hap-4A-2 allele in elite Chinese germplasms is already about 75%.
Conclusions
The gene sequence of TaSDIR1-4A is conserved, and two nucleotide variations were identified in its promoter re- gion, namely a 2-bp InDel and a SNP. Based on the SNP at position 395 (G/A), a functional marker was developed to distinguish the two haplotypes. Association analysis demonstrated that Hap-4A-2 was the favorable haplotype for high 1000-grain weight (TGW), with genotypes pos- sessing it having lower TaSDIR1-4A expression and higher TGW than those possessing Hap-4A-1. Phenotypic analysis of gene-silenced wheat and transgenic Arabidopsis demon- strated that TaSDIR1-4A is a negative regulator for grain size. The functional marker that we have developed for Hap-4A-2 should be helpful for future molecular breeding in wheat.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Alignment of sequences of plant SDIR1 homologs and phylogenic analysis.
Fig. S2. Sequence alignment of TaSDIR1-4A between 32 wheat accessions.
Fig. S3. Chromosome location and genetic mapping of
![]() TaSDIR1-4A.
TaSDIR1-4A.
Fig. S4. Chlorotic stripe mosaic symptoms of BSMV-infected plants and the specificity of knock-down.
Fig. S5. Semi-quantitative PCR for TaSDIR1-4A expression levels in Arabidopsis.
Fig. S6. Expression and purification of the TaERF-BD protein.
Fig. S7. Expression patterns of TaERF3 in different tissues of wheat at different stages of growth.
Fig. S8. A working model of the role of TaSDIR1-4A in determining 1000-grain weight.
Table S1. List of the 32 accessions of the wheat population used for the detection of nucleotide polymorphisms.
Table S2. Primers used in this study.
Table S3. Accessions possessing the different haplotypes of TaSDIR1-4A that were randomly selected from Populations 1 and 2.
Table S4. The 262 accessions of Population 2 and their
TaSDIR1-4A haplotypes.
Acknowledgments
We thank Professor Robert A. McIntosh (Plant Breeding Institute, University of Sydney, Australia) for suggested revisions to the manu- script. We also thank Professor Zengyan Zhang and Dr Xiuliang Zhu (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing the VIGS vectors and for guidance in their use. This study was supported by the National Key R&D Program of China (2017YFD0300202), and the National Natural Science Foundation of China (31461143024).
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.
References
Aya K, Hobo T, Sato-Izawa K, Ueguchi-Tanaka M, Kitano H, Matsuoka M. 2014. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant & Cell Physiology 55, 897–912.
Bednarek J, Boulaflous A, Girousse C, Ravel C, Tassy C, Barret P, Bouzidi MF, Mouzeyar S. 2012. Down-regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. Journal of Experimental Botany 63, 5945–5955.
Boyer JS. 1982. Plant productivity and environment. Science 218, 443–448.
Cao SH, Xu DA, Hanif M, Xia XC, He ZH. 2020. Genetic architecture underpinning yield component traits in wheat. Theoretical and Applied Genetics 133, 1811–1823.
![]()
Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743.
Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M. 2006. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Current Biology 16, 272–279.
Dong H, Dumenil J, Lu FH, et al. 2017. Ubiquitylation activates a pep- tidase that promotes cleavage and destabilization of its activating E3 lig- ases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis. Genes and Development 31, 197–208.
Du L, Li N, Chen LL, Xu YX, Li Y, Zhang YY, Li CY, Li YH. 2014. The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. The Plant Cell 26, 665–677.
Gao T, Wu Y, Zhang Y, Liu L, Ning Y, Wang D, Tong H, Chen S, Chu C, Xie Q. 2011. OsSDIR1 overexpression greatly improves drought tolerance in transgenic rice. Plant Molecular Biology 76, 145–156.
Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C. 2010. Food security: the challenge of feeding 9 billion people. Science 327, 812–818.
Hao C, Wang L, Ge H, Dong Y, Zhang X. 2011. Genetic diversity and linkage disequilibrium in Chinese bread wheat (Triticum aestivum L.) re- vealed by SSR markers. PloS ONE 6, e17279.
Hong Y, Chen L, Du LP, Su Z, Wang J, Ye X, Qi L, Zhang Z. 2014. Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Functional and Integrative Genomics 14, 341–349.
Jaiswal V, Gahlaut V, Mathur S, Agarwal P, Khandelwal MK, Khurana JP, Tyagi AK, Balyan HS, Gupta PK. 2015. Identification of novel SNP in promoter sequence of TaGW2-6A associated with grain weight and other agronomic traits in wheat (Triticum aestivum L.). PloS ONE 10, e0129400.
Kagale S, Links MG, Rozwadowski K. 2010. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic re- pression motif-containing transcriptional regulators in Arabidopsis. Plant Physiology 152, 1109–1134.
Kelley DR. 2018. E3 ubiquitin ligases: key regulators of hormone signaling in plants. Molecular & Cellular Proteomics 17, 1047–1054.
Li HW, Tong YP, Li B, Jing RL, Lu CM, Li ZS. 2010a. Genetic analysis of tolerance to photo-oxidative stress induced by high light in winter wheat (Triticum aestivum L.). Journal of Genetics and Genomics 37, 399–412.
Li L, Mao X, Wang J, Chang X, Reynolds M, Jing R. 2019a. Genetic dissection of drought and heat-responsive agronomic traits in wheat. Plant Cell and Environment 42, 2540–2553.
Li N, Li Y. 2016. Signaling pathways of seed size control in plants. Current Opinion in Plant Biology 33, 23–32.
Li N, Xu R, Duan P, Li, Y. 2018. Control of grain size in rice. Plant Reproduction 31, 237–251.
Li N, Xu R, Li Y. 2019b. Molecular networks of seed size control in plants. Annual Review of Plant Biology 70, 435–463.
Li Q, Li L, Yang XH, Warburton ML, Bai GH, Dai JR, Li JS, Yan JB. 2010b. Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biology 10, 143–157.
Li SP, Wang CS, Chang XP, Jing, RL. 2012a. Genetic dissection of devel- opmental behavior of grain weight in wheat under diverse temperature and water regimes. Genetica 140, 393–405.
Li W, Zhang B, Zhang J, Chang X, Li R, Jing R. 2012b. Exploring elite alleles for chlorophyll content of flag leaf in natural population of wheat by association analysis. Acta Agronomica Sinica 38, 962–970.
Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. 2007. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318, 1302–1305.
Liu H, Li HF, Hao CY, Wang K, Wang YM, Qin L, An DG, Li T, Zhang, XY. 2020. TaDA1, a conserved negative regulator of kernel size, has an addi- tive effect with TaGW2 in common wheat (Triticum aestivum L.). Plant Biotechnology Journal 18, 1330–1342.
Liu J, Xia Z, Wang M, Zhang X, Yang T, Wu J. 2013. Overexpression of a maize E3 ubiquitin ligase gene enhances drought tolerance through regu- lating stomatal aperture and antioxidant system in transgenic tobacco. Plant Physiology and Biochemistry 73, 114–120.
Müller M, Munné-Bosch S. 2015. Ethylene response factors: a key regu- latory hub in hormone and stress signaling. Plant Physiology 169, 32–41.
Nadolska-Orczyk A, Rajchel IK, Orczyk W, Gasparis S. 2017. Major genes determining yield-related traits in wheat and barley. Theoretical and Applied Genetics 130, 1081–1098.
Oh SJ, Kim YS, Kwon CW, Park HK, Jeong JS, Kim JK. 2009. Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiology 150, 1368–1379.
Oh TR, Kim JH, Cho SK, Ryu MY, Yang SW, Kim WT. 2017. AtAIRP2
![]() E3 ligase affects ABA and high-salinity responses by stimulating its ATP1/ SDIRIP1 substrate turnover. Plant Physiology 174, 2515–2531.
E3 ligase affects ABA and high-salinity responses by stimulating its ATP1/ SDIRIP1 substrate turnover. Plant Physiology 174, 2515–2531.
Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. 2001. Repression domains of class II ERF transcriptional repressors share an es- sential motif for active repression. The Plant Cell 13, 1959–1968.
Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ. 2005. Control of seed mass by APETALA2. Proceedings of the National Academy of Sciences, USA 102, 3123–3128.
Schmidt R, Schippers JH, Mieulet D, Watanabe M, Hoefgen R, Guiderdoni E, Mueller-Roeber B. 2014. SALT-RESPONSIVE ERF1 is a negative regulator of grain filling and gibberellin-mediated seedling estab- lishment in rice. Molecular Plant 7, 404–421.
Simmonds J, Scott P, Brinton J, Mestre TC, Bush M, Del Blanco A, Dubcovsky J, Uauy C. 2016. A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theoretical and Applied Genetics 129, 1099–1112.
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. 2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nature Genetics 39, 623–630.
Su Z, Hao C, Wang L, Dong Y, Zhang X. 2011. Identification and devel- opment of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theoretical and Applied Genetics 122, 211–223.
Sun J, Sun Y, Ahmed RI, Ren A, Xie MM. 2019. Research progress on plant RING-finger proteins. Genes 10, 973–988.
Tak H, Mhatre M. 2013. Molecular characterization of VvSDIR1 from Vitis vinifera and its functional analysis by heterologous expression in Nicotiana tabacum. Protoplasma 250, 565–576.
Tilman D, Balzer C, Hill J, Befort BL. 2011. Global food demand and the sustainable intensification of agriculture. Proceedings of the National Academy of Sciences, USA 108, 20260–20264.
Wang J, Li Q, Mao X, Li A, Jing R. 2016a. Wheat transcription factor TaAREB3 participates in drought and freezing tolerances in Arabidopsis. International Journal of Biological Sciences 12, 257–269.
Wang J, Wang R, Mao X, Li L, Chang X, Zhang X, Jing R. 2019. TaARF4 genes are linked to root growth and plant height in wheat. Annals of Botany 124, 903–915.
Wang JL, Tang MQ, Chen S, et al. 2017. Down-regulation of BnDA1, whose gene locus is associated with the seeds weight, improves the seeds weight and organ size in Brassica napus. Plant Biotechnology Journal 15, 1024–1033.
Wang Q, Qi W, Wang Y, Sun F, Qian X, Luo X, Yang J. 2011. Isolation and identification of an AP2/ERF factor that binds an allelic cis-element of rice gene LRK6. Genetics Research 93, 319–332.
Wang R, Wang J, Mao X, Chang X, Liu H, Jing R. 2018. Cloning and functional analysis of a RING-type E3 ubiquitin ligase gene TaSDIR1-D in wheat. Journal of Plant Genetic Resources 19, 951–958. doi:10.13430/j. cnki.jpgr.20180309002. [In Chinese, with English abstract]
Wang X, Wang H, Liu S, Ferjani A, Li J, Yan J, Yang X, Qin F. 2016b. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nature Genetics 48, 1233–1241.
Wei X, Xu H, Rong W, Ye X, Zhang Z. 2019. Constitutive expression of a stabilized transcription factor group VII ethylene response factor enhances waterlogging tolerance in wheat without penalizing grain yield. Plant, Cell & Environment 42, 1471–1485.
Xia T, Li N, Dumenil J, Li J, Kamenski A, Bevan MW, Gao F, Li Y. 2013a. The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. The Plant Cell 25, 3347–3359.
Xia Z, Liu Q, Wu J, Ding J. 2012. ZmRFP1, the putative ortholog of SDIR1, encodes a RING-H2 E3 ubiquitin ligase and responds to drought stress in an ABA-dependent manner in maize. Gene 495, 146–153.
Xia Z, Su X, Liu J, Wang M. 2013b. The RING-H2 finger gene 1 (RHF1) encodes an E3 ubiquitin ligase and participates in drought stress response in Nicotiana tabacum. Genetica 141, 11–21.
Xie G, Li Z, Ran Q, Wang H, Zhang J. 2018. Over-expression of mutated ZmDA1 or ZmDAR1 gene improves maize kernel yield by enhancing starch synthesis. Plant Biotechnology Journal 16, 234–244.
Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH. 2002. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170.
Xu FQ, Xue HW. 2019. The ubiquitin-proteasome system in plant re- sponses to environments. Plant, Cell & Environment 42, 2931–2944.
Xu JJ, Zhang XF, Xue HW. 2016. Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. Journal of Experimental Botany 67, 6399–6411.
Yang DL, Jing RL, Chang XP, Li W. 2007. Identification of quantitative trait loci and environmental interactions for accumulation and remobilization of water-soluble carbohydrates in wheat (Triticum aestivum L.) stems. Genetics 176, 571–584.
Zhang B, Shi W, Li W, Chang X, Jing R. 2013. Efficacy of pyramiding elite alleles for dynamic development of plant height in common wheat. Molecular Breeding 32, 327–338.
Zhang B, Xu W, Liu X, Mao X, Li A, Wang J, Chang X, Zhang X, Jing R.
2017. Functional conservation and divergence among homoeologs of
TaSPL20 and TaSPL21, two SBP-box genes governing yield-related traits in hexaploid wheat. Plant Physiology 174, 1177–1191.
Zhang H, Cui F, Wu Y, Lou L, Liu L, Tian M, Ning Y, Shu K, Tang S, Xie Q. 2015. The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1- INTERACTING PROTEIN1 for degradation to modulate the salt stress re- sponse and ABA signaling in Arabidopsis. The Plant Cell 27, 214–227.
![]() Zhang XY, Li CW, Wang LF, Wang HM, You GX, Dong YS. 2002. An estimation of the minimum number of SSR alleles needed to reveal genetic relationships in wheat varieties. I. Information from large-scale planted var- ieties and cornerstone breeding parents in Chinese wheat improvement and production. Theoretical and Applied Genetics 106, 112–117.
Zhang XY, Li CW, Wang LF, Wang HM, You GX, Dong YS. 2002. An estimation of the minimum number of SSR alleles needed to reveal genetic relationships in wheat varieties. I. Information from large-scale planted var- ieties and cornerstone breeding parents in Chinese wheat improvement and production. Theoretical and Applied Genetics 106, 112–117.
Zhang Y, Li D, Zhang D, et al. 2018. Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. The Plant Journal 94, 857–866.
Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. 2007. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. The Plant Cell 19, 1912–1929.
Zhang YY, Li Y, Gao T, et al. 2008. Arabidopsis SDIR1 enhances drought tolerance in crop plants. Bioscience, Biotechnology, and Biochemistry 72, 2251–2254.
Zuo J, Li J. 2014. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annual Review of Genetics 48, 99–118.
